I don't have any data to support this next claim, but I'm fairly sure that Excel is the most common way to store, manipulate, and yes(!), even pass data around. This is why it's not uncommon to find yourself reading Excel in Python. I recently needed to, so I tested and benchmarked several ways of reading Excel files in Python.
In this article I compare several ways to read Excel from Python.
Table of Contents
What are we Testing?
To compare ways to read Excel files with Python, we first need to establish what to measure, and how.
We start by creating a 25MB Excel file containing 500K rows with various column types:
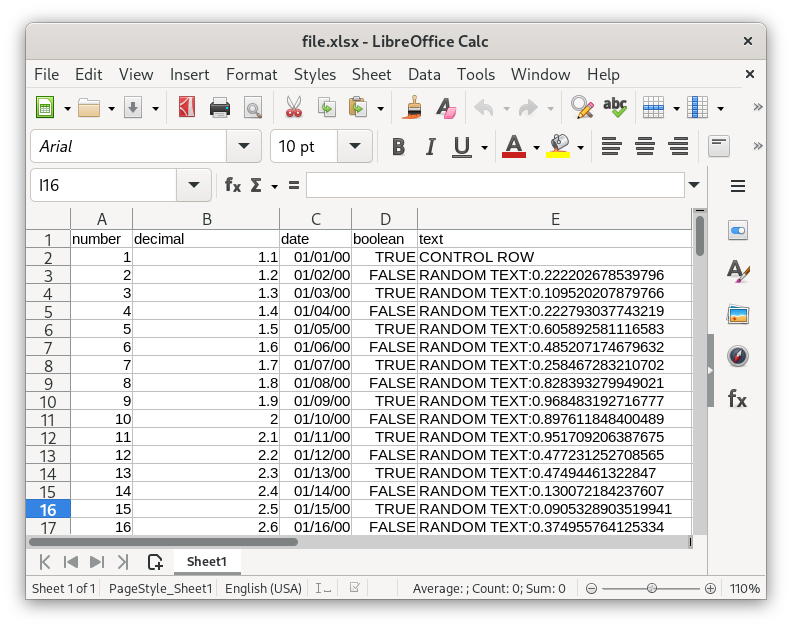
Excel supports both the xls and the xlsx file formats. We'll use the newer format xlsx.
For the benchmarks, we'll implement functions to import data from Excel and return an Iterator of dicts:
def iter_excel(file: IO[bytes]) -> Iterator[dict[str, object]]:
# TODO...
We return an Iterator to allow consumers to process the file row by row. This can potentially reduce the memory footprint by not storing the entire file in memory as we process it. As we'll see in the benchmarks, this is not always possible.
To produce a "clean" timing we iterate the generator without actually doing any processing:
for row in iter_excel(file):
pass
This will cause the generator to fully evaluate with minimal performance or memory overhead.
Speed
The most obvious thing to measure is time, and the most accurate way to measure time in Python for performance purposes is using time.perf_counter:
import time
start = time.perf_counter()
for row in iter_excel(file): pass
elapsed = time.perf_counter() - start
We start the timer, iterate the entire generator and calculate the elapsed time.
Types
Some formats such as parquet and avro are known for being self-describing, keeping the schema inside the file, while other formats such as CSV are notorious for not keeping any information about the data they store.
Excel can be seen as a format that does store type information about its content - there are Date cells, Number cells, Decimal cells and others, so when loading data from Excel, it can be useful to receive the data in its intended type. This is especially useful for types such as date, where the format may be unclear or unknown, or strings that hold digits such as phone numbers or zipcodes. In these situations, trying to sniff the type can cause incorrect results (due to trimming leading zeros, assuming incorrect format and so on).
To be fair, some may argue that when loading data into your system you should have knowledge about its schema, so preserving types may not be a strict requirement for some.
Correctness
To test the correctness of the import process, we include a control row at the beginning of the Excel file. We'll use the control row as reference to make sure the data is imported correctly:
# Test correctness of imported data using a control row
for key, expected_value in (
('number', 1),
('decimal', 1.1),
('date', datetime.date(2000, 1, 1)),
('boolean', True),
('text', 'CONTROL ROW'),
):
try:
value = control_row[key]
except KeyError:
print(f'🔴 "{key}" missing')
continue
if type(expected_value) != type(value):
print(f'🔴 "{key}" expected type "{type(expected_value)}" received type "{type(value)}"')
elif expected_value != value:
print(f'🔴 "{key}" expected value "{expected_value}" received "{value}"')
else:
print(f'🟢 "{key}"')
We'll run this test after each benchmark to make sure that all of the expected keys exist in the control row, and that the types and values are as we expect.
Reading Excel in Python
We now have a sample file, a way to test the contents and we've defined what to measure - we are ready to import some data!
Reading Excel using Pandas
Pandas, the data analysis library for Python, is the go-to for just about anything related to data in Python, so it's a good place to start.
Read an Excel file using pandas:
import pandas
def iter_excel_pandas(file: IO[bytes]) -> Iterator[dict[str, object]]:
yield from pandas.read_excel(file).to_dict('records')
Just two commands chained together to get a list of dictionaries from an Excel file. This is what a single row from the result looks like:
>>> with open('file.xlsx', 'rb') as f:
... rows = iter_excel_pandas(f)
... row = next(rows)
... print(row)
...
{'boolean': True,
'date': Timestamp('2000-01-01 00:00:00'),
'decimal': 1.1,
'number': 1,
'text': 'CONTROL ROW'}
At a quick glance we can see the date is not a datetime.date but a pandas Timestamp. The rest looks OK. If the Timestamp is an issue and you insist on datetime.date, you can provide a converter function to read_excel:
import pandas
def iter_excel_pandas(file: IO[bytes]) -> Iterator[dict[str, object]]:
yield from pandas.read_excel(file, converters={
'date': lambda ts: ts.date(),
}).to_dict('records')
The converter accepts a pandas Timestamp and converts is to a datetime.date. This is the control row with the custom converter:
{
'number': 1,
'decimal': 1.1,
'date': datetime.date(2000, 1, 1),
'boolean': True,
'text': 'CONTROL ROW',
}
If you use pandas to read data from Excel it's not unreasonable to assume you also want to continue your analysis with pandas, so we'll accept the Timestamp as a valid type for our benchmark.
Next, run the benchmark on the large Excel file:
iter_excel_pandas
elapsed 32.98058952600695
🟢 "number"
🟢 "decimal"
🔴 "date" expected type "<class 'datetime.date'>" received type "<class 'pandas._libs.tslibs.timestamps.Timestamp'>"
🟢 "boolean"
🟢 "text"
The import took ~32s to complete. The type of the date field is a pandas Timestamp and not datetime.date, but that's OK.
Reading Excel using Tablib
Tablib is one of the most popular libraries in Python for importing and exporting data in various formats. It was originally developed by the creator of the popular requests library, and therefor characterized by a similar focus on developer experience and ergonomics.
To install Tablib, execute the following command:
$ pip install tablib
Read an Excel file using tablib:
import tablib
def iter_excel_tablib(file: IO[bytes]) -> Iterator[dict[str, object]]:
yield from tablib.Dataset().load(file).dict
Just a single line of code and the library does all the heavy lifting.
Before we go on to execute the benchmark, this is how the first row of the results looks like:
>>> with open('file.xlsx', 'rb') as f:
... rows = iter_excel_tablib(f)
... row = next(rows)
... print(row)
...
OrderedDict([('number', 1),
('decimal', 1.1),
('date', datetime.datetime(2000, 1, 1, 0, 0)),
('boolean', True),
('text', 'CONTROL ROW')])
OrderedDict is a subclass of a Python dict with some additional methods to rearrange the dictionary order. It's defined in the built-in collections module and it is what tablib returns when you ask for a dict. Since OrderedDict is a subclass of dict and it's defined in a built-in module, we don't mind and consider it just fine for our purposes.
Now for the benchmark on the large Excel file:
iter_excel_tablib
elapsed 28.526969947852194
🟢 "number"
🟢 "decimal"
🔴 "date" expected type "<class 'datetime.date'>" received type "<class 'datetime.datetime'>"
🟢 "boolean"
🟢 "text"
Import using tablib took 28s, faster than pandas (32s). The date cell was returned as a datetime.datetime instead of a datetime.date, not unreasonable.
Let's see if we can bring this timing down even further.
Reading Excel using Openpyxl
Openpyxl is a library for reading and writing Excel files in Python. Unlike Tablib, Openpyxl is dedicated just to Excel and does not support any other file types. In fact, both tablib and pandas use Openpyxl under the hood when reading xlsx files. Perhaps this specialization will result in better performance.
To install openpyxl, execute the following command:
$ pip install openpyxl
Read an Excel file using openpyxl:
import openpyxl
def iter_excel_openpyxl(file: IO[bytes]) -> Iterator[dict[str, object]]:
workbook = openpyxl.load_workbook(file)
rows = workbook.active.rows
headers = [str(cell.value) for cell in next(rows)]
for row in rows:
yield dict(zip(headers, (cell.value for cell in row)))
This time we have to write a bit more code, so let's break it down:
-
Load a workbook from the open file: The function
load_workbooksupports both a file path and a readable stream. In our case we operate on an open file. -
Get the active sheet: An Excel file can contain multiple sheets and we can choose which one to read. In our case we only have one sheet.
-
Construct a list of headers: The first row in the Excel file includes the headers. To use these header as keys for our dictionary we read the first row and produce the list of headers.
-
Return the results: Iterate the rows and construct a dictionary for each row using the headers and the cell values.
openpyxluses a Cell type that includes both the value and some metadata. This can be useful for other purposes, but we only need the values. To access a cell's value we usecell.value.
This is what the first row of the results looks like:
>>> with open('file.xlsx', 'rb') as f:
... rows = iter_excel_openpyxl(f)
... row = next(rows)
... print(row)
{'boolean': True,
'date': datetime.datetime(2000, 1, 1, 0, 0),
'decimal': 1.1,
'number': 1,
'text': 'CONTROL ROW'}
Looks promising! Run the benchmark on the large file:
iter_excel_openpyxl
elapsed 35.62
🟢 "number"
🟢 "decimal"
🔴 "date" expected type "<class 'datetime.date'>" received type "<class 'datetime.datetime'>"
🟢 "boolean"
🟢 "text"
Importing the large Excel file using openpyxl took ~35s, longer then the Tablib (28s) and pandas (32s).
A quick search at the documentation revealed a promising section titled "performance". In this section, openpyxl describes "optimized modes" to speed things up when only reading or writing a file:
import openpyxl
def iter_excel_openpyxl(file: IO[bytes]) -> Iterator[dict[str, object]]:
workbook = openpyxl.load_workbook(file, read_only=True)
rows = workbook.active.rows
headers = [str(cell.value) for cell in next(rows)]
for row in rows:
yield dict(zip(headers, (cell.value for cell in row)))
The worksheet in now loaded in "read only" mode. Since we only want to read the contents and not write, this is acceptable. Let's run the benchmark again and see if it affected the results:
iter_excel_openpyxl
elapsed 24.79
🟢 "number"
🟢 "decimal"
🔴 "date" expected type "<class 'datetime.date'>" received type "<class 'datetime.datetime'>"
🟢 "boolean"
🟢 "text"
Opening the file in "read only" mode brings the timing down from 35s to 24s - faster than tablib (28s) and pandas (32s).
Reading Excel using LibreOffice
We have now exhausted the traditional and obvious ways to import Excel into Python. We used the top designated libraries and got decent results. It's now the time to think outside the box.
LibreOffice is a free and open source alternative to the other office suite. LibreOffice can process both xls and xlsx files and also happens to include a headless mode with some useful command line options:
$ libreoffice --help
LibreOffice 7.5.8.2 50(Build:2)
Usage: soffice [argument...]
argument - switches, switch parameters and document URIs (filenames).
...
One of the LibreOffice command line options is to convert files between different formats. For example, we can use libreoffice to convert an xlsx file to a csv file:
$ libreoffice --headless --convert-to csv --outdir . file.xlsx
convert file.xlsx -> file.csv using filter: Text - txt - csv (StarCalc)
$ head file.csv
number,decimal,date,boolean,text
1,1.1,01/01/2000,TRUE,CONTROL ROW
2,1.2,01/02/2000,FALSE,RANDOM TEXT:0.716658989024692
3,1.3,01/03/2000,TRUE,RANDOM TEXT:0.966075283958641
Nice! Let's stich it together using Python. We'll first convert the xlsx file to CSV and then import the CSV into Python:
import subprocess, tempfile, csv
def iter_excel_libreoffice(file: IO[bytes]) -> Iterator[dict[str, object]]:
with tempfile.TemporaryDirectory(prefix='excelbenchmark') as tempdir:
subprocess.run([
'libreoffice', '--headless', '--convert-to', 'csv',
'--outdir', tempdir, file.name,
])
with open(f'{tempdir}/{file.name.rsplit(".")[0]}.csv', 'r') as f:
rows = csv.reader(f)
headers = list(map(str, next(rows)))
for row in rows:
yield dict(zip(headers, row))
Let's break it down:
-
Create a temporary directory for storing our CSV file: Use the built-in
tempfilemodule to create a temporary directory that will cleanup automatically when we are done. Ideally, we would want to convert a specific file into a file-like-object in memory, but thelibreofficecommand line does not provide any way of converting into a specific file, only to a directory. -
Convert to CSV using the
libreofficecommand line: Use the built-insubprocessmodule to execute an OS command. -
Read the generated CSV: Open the newly created CSV file, parse it using the build-in
csvmodule and produce dicts.
This is what the first row of the results looks like:
>>> with open('file.xlsx', 'rb') as f:
... rows = iter_excel_libreoffice(f)
... row = next(rows)
... print(row)
{'number': '1',
'decimal': '1.1',
'date': '01/01/2000',
'boolean': 'TRUE',
'text': 'CONTROL ROW'}
We immediately notice that we lost all the type information - all values are strings.
Let's run the benchmark to see if it's worth it:
iter_excel_libreoffice
convert file.xlsx -> file.csv using filter : Text - txt - csv (StarCalc)
elapsed 15.279242266900837
🔴 "number" expected type "<class 'int'>" received type "<class 'str'>"
🔴 "decimal" expected type "<class 'float'>" received type "<class 'str'>"
🔴 "date" expected type "<class 'datetime.date'>" received type "<class 'str'>"
🔴 "boolean" expected type "<class 'bool'>" received type "<class 'str'>"
🟢 "text"
To be honest, this was faster than I had anticipated! Using LibreOffice to convert the file to CSV and then loading it took only 15s - faster than pandas (35s), tablib (28s) and openpyxl (24s).
We did lose the type information when we converted the file to CSV and if we had to also convert the types it will most likely take a bit more time (serialization can be slow you know). But overall, not a bad option!
Reading Excel using DuckDB
If we're already down the path of using external tools, why not give the new kid on the block a chance at competing.
DuckDB is an "in-process SQL OLAP database management system". This description does not make it immediately clear why DuckDB can be useful in this case, but it is. DuckDB is very good at moving data around and converting between formats.
To install the DuckDB Python API execute the following command:
$ pip install duckdb
Read an Excel file using duckdb in Python:
import duckdb
def iter_excel_duckdb(file: IO[bytes]) -> Iterator[dict[str, object]]:
duckdb.install_extension('spatial')
duckdb.load_extension('spatial')
rows = duckdb.sql(f"""
SELECT * FROM st_read(
'{file.name}',
open_options=['HEADERS=FORCE', 'FIELD_TYPES=AUTO'])
""")
while row := rows.fetchone():
yield dict(zip(rows.columns, row))
Let's break it down:
-
Install and load the
spatialextension: To import data from Excel usingduckdbyou need install thespatialextension. This is a bit strange becausespatialis used for geo manipulations, but that's what it wants. -
Query the file: When executing queries directly using the
duckdbglobal variable it will use an in-memory database by default, similar to usingsqlitewith the:memory:option. To actually import the Excel file, we use thest_readfunction with the path to the file as first argument. In the function options, we set the first row as headers, and activate the option to automatically detect types (this is also the default). -
Construct the result: Iterate the rows and construct dicts using the list of headers and values of each row.
This is what the first row looks like using DuckDB to import the Excel file:
>>> with open('file.xlsx', 'rb') as f:
... rows = iter_excel_duckdb(f)
... row = next(rows)
... print(row)
{'boolean': True,
'date': datetime.date(2000, 1, 1),
'decimal': 1.1,
'number': 1,
'text': 'CONTROL ROW'}
Now that we have our process to read an Excel file using DuckDB to Python, let's see how it performs:
iter_excel_duckdb
elapsed 11.36
🟢 "number"
🟢 "decimal"
🟢 "date"
🟢 "boolean"
🟢 "text"
First of all, we have a winner with the types! DuckDB was able to correctly detect all the types. Additionally, DuckDB clocked at only 11s, which brings us closer to a single digit timing!
One thing that bothered me with this implementation was that despite my best efforts, I was unable to use a parameter for the name of the file using the duckdb.sql function. Using string concatenation to generate SQL is dangerous, prone to injection and should be avoided when possible.
In one of my attempts to resolve this, I tried to use duckdb.execute instead of duckdb.sql, which seemed to accept parameters in this case:
import duckdb
def iter_excel_duckdb_execute(file: IO[bytes]) -> Iterator[dict[str, object]]:
duckdb.install_extension('spatial')
duckdb.load_extension('spatial')
conn = duckdb.execute(
"SELECT * FROM st_read(?, open_options=['HEADERS=FORCE', 'FIELD_TYPES=AUTO'])",
[file.name],
)
headers = [header for header, *rest in conn.description]
while row := conn.fetchone():
yield dict(zip(headers, row))
There are two main differences here:
-
Use
duckdb.executeinstead ofduckdb.sql: UsingexecuteI was able to provide the file name as parameter rather than using string concatenation. This is safer. -
Construct the headers: According to the API reference,
duckdb.sqlreturns aDuckDBPyRelationwhileduckdb.executereturns aDuckDBPyConnection. To produce a list of headers from the connection object, I was unable to access.columnslike before, so I had to look at thedescriptionproperty of the connection, which I imagine describes the current result set.
Running the benchmark using the new function yielded some interesting results:
iter_excel_duckdb_execute
elapsed 5.73
🔴 "number" expected type "<class 'int'>" received type "<class 'str'>"
🔴 "decimal" expected type "<class 'float'>" received type "<class 'str'>"
🔴 "date" expected type "<class 'datetime.date'>" received type "<class 'str'>"
🔴 "boolean" expected type "<class 'bool'>" received type "<class 'str'>"
🟢 "text"
Using execute we gobbled the file in just 5.7s - that's twice as fast as the last attempt, but we lost the types. Without much knowledge and experience using DuckDB I can only assume constructing the relation and casting to the correct types incurs some overhead.
Before we move on to other options, let's check if pre-loading and installing the extensions makes any significant difference:
import duckdb
+duckdb.install_extension('spatial')
+duckdb.load_extension('spatial')
+
def iter_excel_duckdb_execute(file: IO[bytes]) -> Iterator[dict[str, object]]:
- duckdb.install_extension('spatial')
- duckdb.load_extension('spatial')
rows = duckdb.execute(
"SELECT * FROM st_read(?, open_options=['HEADERS=FORCE', 'FIELD_TYPES=AUTO'])",
Executing the function several times:
iter_excel_duckdb_execute
elapsed 5.28
elapsed 5.69
elapsed 5.28
Pre-loading the extensions did not have a significant effect on the timing.
Let's see if removing the automatic type detection has any effect:
duckdb.load_extension('spatial')
def iter_excel_duckdb_execute(file: IO[bytes]) -> Iterator[dict[str, object]]:
conn = duckdb.execute(
- "SELECT * FROM st_read(?, open_options=['HEADERS=FORCE', 'FIELD_TYPES=AUTO'])",
+ "SELECT * FROM st_read(?, open_options=['HEADERS=FORCE', 'FIELD_TYPES=STRING'])",
[file.name],
)
headers = [header for header, *rest in conn.description]
Executing the function several times:
iter_excel_duckdb_execute
elapsed 5.80
elapsed 7.21
elapsed 6.45
Removing the automatic type detection also didn't seem to have any significant effect on the timing.
Reading Excel using Calamine
In recent years it seems like every performance problem in Python ends up being solved with another language. As a Python developer, I consider this a true blessing. It means I can keep using the language I'm used to and enjoy the performance benefits of all others!
Calamine is a pure Rust library to read Excel and OpenDocument Spreadsheet files. To install python-calamine, the Python binding for calamine, execute the following command:
$ pip install python-calamine
Read an Excel file using calamine in Python:
import python_calamine
def iter_excel_calamine(file: IO[bytes]) -> Iterator[dict[str, object]]:
workbook = python_calamine.CalamineWorkbook.from_filelike(file) # type: ignore[arg-type]
rows = iter(workbook.get_sheet_by_index(0).to_python())
headers = list(map(str, next(rows)))
for row in rows:
yield dict(zip(headers, row))
Going through the same routine again - load the workbook, pick the sheet, fetch the headers from the first row, iterate the results and construct a dict from every row.
This is what the first row looks like:
>>> with open('file.xlsx', 'rb') as f:
... rows = iter_excel_calamine(f)
... row = next(rows)
... print(row)
{'boolean': True,
'date': datetime.date(2000, 1, 1),
'decimal': 1.1,
'number': 1.0,
'text': 'CONTROL ROW'}
Running the benchmark:
iter_excel_calamine
elapsed 3.58
🔴 "number" expected type "<class 'int'>" received type "<class 'float'>"
🟢 "decimal"
🟢 "date"
🟢 "boolean"
🟢 "text"
That's a big leap! Using python-calamine we processed the entire file in just 3.5s - the fastest so far! The only red dot here is because our integer was interpreted as float - not entirely unreasonable.
After pocking around a bit, the only issue I could find with python-calamine is that it cannot produce results as an iterator. The function CalamineWorkbook.from_filelike will load the entire dataset into memory, which depending on the size of the file, can be an issue. The author of the Python binding library pointed me to this issue in the underlying binding library pyo3, which prevents iteration on Rust structures from Python.
Results Summary
Here is a summary of methods to read Excel files using Python:
| Method | Timing (seconds) | Types | Version |
|---|---|---|---|
| Pandas | 32.98 | Yes | 2.1.3 |
| Tablib | 28.52 | Yes | 3.5.0 |
| Openpyxl | 35.62 | Yes | 3.1.2 |
| Openpyxl (readonly) | 24.79 | Yes | 3.1.2 |
| LibreOffice | 15.27 | No | 7.5.8.2 |
| DuckDB (sql) | 11.36 | Yes | 0.9.2 |
| DuckDB (execute) | 5.73 | No | 0.9.2 |
| Calamine (python-calamine) | 3.58 | Yes | 0.22.1 (0.1.7) |
So which one should you use? it depends... There are a few additional considerations other than speed when choosing a library for working with Excel files in Python:
-
Write capability: we benchmarked ways to read Excel, but sometimes it's necessary to produce Excel files as well. Some of the libraries we benchmarked does not support writing. Calamine for example, cannot write Excel files, only read.
-
Additional formats: A system may require loading and producing files in other formats other than Excel. Some libraries, such as pandas and Tablib support a variety of additional formats, while calamine and openpyxl only support Excel.
Source
The source code for the benchmarks is available in this repo.
